Dugong - Scientific Linux Container
Dugong is a powerful workstation platform especially designed for scientific computational analysis, mainly of bioinformatics and computational biology, that can be installed in any computational ecosystem, regardless of the operating system and hardware used. In general terms, it’s a Linux system designed specifically for the development of the Open Science concept, based on the reproducibility and replicability of the research.
It’s an implementation built under the MIT license using the Docker platform, an open-source project that automates the implementation of applications within software containers, providing an additional layer of abstraction and automation of operating system-level virtualization on Linux. Docker uses the Linux kernel resource isolation features, such as cgroups and namespaces, as well as the file system AuFS (advanced multi layered unification filesystem) to allow independent containers to run in a single Linux instance, avoiding an overload of initialization and maintenance of virtual machines. The Dugong project enables efficient creation of reusable containers for bioinformatics data, making the analysis environment and all computational results reproducible by the scientific community.
Dugong is based on version 16.04 of Ubuntu Image, the only Docker system focused on bioinformatics that provides users with a complete graphical user interface (GUI) XFCE4, independent of the host, which should facilitate user interaction with different computing ecosystems, as well as the installation and use of different bioinformatics software. For the distribution of bioinformatics software, two large open source projects have been integrated Dugong: LinuxBrew and BioConda, totaling over 3,500 bioinformatics packages and their respective dependencies. Dugong is the only system focused on bioinformatics that offers Anaconda Navigator, a graphical package manager for Conda and the BioConda repository, allowing the installation of packages and tools through its graphical interface. We also added, as an alternative, the BioLinux and CloudBioLinux project repositories for the installation of DEB packages through the APT command.
Thinking about the replicability and reproducibility of different lines of research that benefit from bioinformatics analysis, Dugong is the only tool of its kind that provides Jupyter Notebook installed by default on all their versions.
All of these features, among others, make Dugong one of the most complete Docker desktop environments available today, and allow any user with minimal computing knowledge to run a Dugong container in high performance computing (HPC) environments, Virtual Private Server (VPS), as well as in private, public, or hybrid cloud computing environments of different commercial service providers.
The Dugong project is publicly committed to the principles of open source software development, where people are encouraged to use free software, study how they function, improve and distribute them freely.
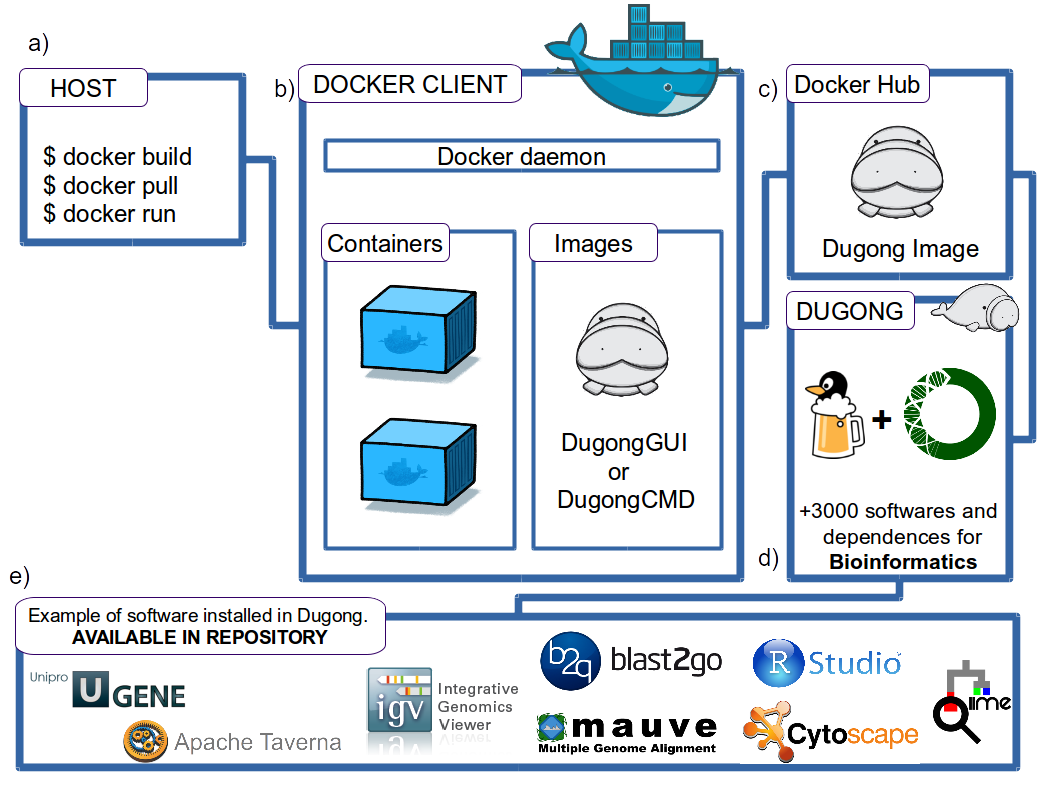
Through the simple Docker Client commands, the user can download the Dugong images (a), and install and manage the containers generated using the Docker Daemon on the host computer (b). The Dugong image is currently available in Docker Hub (c), containing more than 3000 bioinformatics software and its dependencies thanks to the integration of the BioConda and LinuxBrew projects (d).
Among the software that can be installed in Dugong, and enjoy its graphical interface XFCE4, are: Blast2GO, UGene, Apache Taverna, Integrative Genomics Viewer (IGV), Mauve, RStudio, Cytoscape, QIIME (e), among others. Some of these software are already functional in a Dugong image in our GitHub repository.
Test Dugong NOW!
To test the operation of Dugong and demonstrate the ease of use and the possibility of using the service in different operating systems and platforms we provide some installation templates on dply.co’s servers.
dply.co allows the creation of an absolutely free server for two hours, and can be used for a longer time for payment. If you choose not to buy more time for the server, it is deleted automatically after 2 hours without any charge.
dply.co servers are hosted in the DigitalOcean cloud and have the following specifications:
- 1 CPU
- 512MB RAM
- 20GB Disk
We currently offer the following operating systems: CentOS 6, CentOS 7, Debian 7, Debian 8, Fedora 24, Fedora 25, Ubuntu 14.04, Ubuntu 16.04.
Test the Dugong on a machine with Ubuntu 16.04 by clicking the button:
Learn more about Dply and other ways to test Dugong on other operating systems by clicking here.
MENU
- Dugong flavours
- Installing Bioinformatics Software
- Reproducibility and Replicability
- Deploy Dugong Image
- Deploy Dugong Image by graphical interface
- Access Dugong Container
- Extending or adapting the Dugong image
- Example of adapted tools, replicability and reproducibility in Dugong
- CirComPara Pipeline
- CirComPara: Simulating reproducibility and replicability in alternative OS
- CirComPara: Simulating reproducibility and replicability using a pipeline in Jupyter Notebook
- CirComPara: Results of simulations
- CirComPara: Quick installation
- Dugong Virtual Box
- Comparison between Dugong and other bioinformatics services
- Author
- How to cite
- Contributing
- MIT License
- Learn More
Dugong flavours [menu]
Today, the Dugong operating system is available in two versions: DugongGUI and DugongCMD.
DugongGUI is the main working environment of the project and our great differential for applications in bioinformatics and computational biology. Based on Ubuntu 16.04, the DugongGUI image has as its main GUI the XFCE4, with a beta version containing the IceWM graphical environment in development.
Already the DugongCMD version is focused on the fast access and availability of bioinformatics tools through an iterative web interface developed in node.js, which can be accessed directly by the browser. This version does not have a GUI installed by default, but allows X11 forwarding to the host machine.
An important feature of Dugong is that both versions provide system access through the SSH protocol and the exchange of files through RSYNC and SCP. Also providing BaseMount, an opensource application that allows you to mount the BaseSpace Sequence Hub in Dugong, allowing the user to navigate in projects, samples, runs and results of applications and interact directly with associated files.
Below we describe the tools available in each version of Dugong:
DugongGUI Version [menu]
This is the preinstalled software suite that enables better use of DugongGUI:
- Xfce4 (translated as four individual letters) is a free and open source desktop environment for Unix platforms such as Linux, Solaris and BSD. As a fast, easy-to-use open-source workbench, it was selected as the official GUI for the Dugong project.
- IceWM is a window manager designed for lightweight simplicity and ease of use, but not focused on customizability. It looks similar to Windows 95. In beta testing at DugongGUI.
Access as graphical interfaces available in DugongGUI can be performed through two options:
-
VNC: a graphical desktop sharing system that uses the Remote Frame Buffer (RFB) protocol to remotely control another computer. It transmits the keyboard and mouse events from one computer to another, transmitting as graphical screen updates back in the other direction, over a network.
-
noVNC: is an HTML5-based remote desktop web client that can communicate with a remote VNC server via Web Sockets. The noVNC architecture is composed of 6 main components:
- VNC / RFB Deployment Core: This component encapsulates all the intelligence of the RFB protocol and is the main machine that orchestrates everything else.
- Canvas abstraction: This component provides an abstraction of the HTML5 Canvas APIs. It also has the Canvas detection functionality to handle browsers that do not have the full implementation of HTML5.
- User Interface: Any interaction with the HTML DOM (except Canvas) is encapsulated in this component. It renders page controls such as the connect / disconnect button, settings, and the status of responses. This is the main component that allows easy integration of noVNC with Dugong and other tools.
- Utilities: This contains several generic routines and extensions used by noVNC including: extensions to Javascript arrays to make them more useful as queues, event management between browsers, debugging / logging.
- WebSockets Compatibility: Enables Flash (flex) emulation for browsers that do not have native support for WebSockets, as well as providing encryption support for WebSocktes.
- WebSockets Proxy for TCP: a TCP-to-Websockets proxy script, which implements an initial validation similar to HTTP to establish the connection, and then each packet sent begins with a 0 (zero) byte and ends with a 255 byte. This proxy becomes necessary for translation between WebSockets and the standard TCP socket, while VNC does not implement native support for WebSockets.
The following list shows full features offered by noVNC.
- Supports all modern browsers including those on iOS, Android.
- Supported VNC encodings: raw, copyrect, rre, hextile, tight, tightPNG
- WebSocket SSL/TLS encryption (i.e. “wss://”) support
- 24-bit true color and 8 bit colour mapped
- Supports desktop resize notification/pseudo-encoding
- Local or remote cursor
- Clipboard copy/paste
- Clipping or scrolling modes for large remote screens
More information about noVNC can be found on the project page.
DugongGUI also has a set of preinstalled software packages that allow a better use of the operating system, especially:
- Anaconda Navigator: is a desktop graphical user interface (GUI) that is included in Anaconda or Miniconda, and allows you to launch applications and easily manage Conda and Bioconda packages, environments and channels without using command-line commands.
- Spyder IDE: a powerful interactive development environment for the Python language with advanced editing, interactive testing, debugging and introspection features and a numerical computing environment thanks to the support of IPython (enhanced interactive Python interpreter) and popular Python libraries such as NumPy (linear algebra), SciPy (signal and image processing) or matplotlib (interactive 2D/3D plotting).
- ASCIInema: is a free and open source solution for recording terminal sessions and sharing them on the web.
- Google Chrome: a browser that combines a minimalist design with sophisticated technology to make the web faster, safer and easier.
- MEGASync: a desktop client for MEGA Service Cloud providing easy automatic synchronization of files and folders between our computer and the cloud drive with 50Gb storage.
- MEGAcmd: tool that allows access through the Mega cloud command line and provides a set of powerful commands for a file manipulation.
DugongGUI allows a simple, quick and easy distribution of any bioinformatics application that can be installed on an Ubuntu operating system. It also allows the generated containers to be used for a wide range of purposes, such as training, data analysis, tool implementation, devops, etc. In addition, it allows graphical visualization of results from different computational analyzes, such as graphs and figures of software and statistical tools, among others.
DugongCMD [menu]
This is the preinstalled software suite that enables better use of DugongCMD:
- ASCIInema: is a free and open source solution for recording terminal sessions and sharing them on the web.
- Showterm: is an open source terminal record and upload application that lets you easy to record how-to in your terminal. It will record all your terminal activity in text-base and upload to showterm.io as a video and then generates a link for you to share with your team-mates or embed it in your website as an iframe.
- MEGAcmd: tool that allows access through the Mega cloud command line and provides a set of powerful commands for a file manipulation.
Access to DugongCMD can be done through the following option:
- tty.js: a terminal in your browser using node.js and socket.io. Based on Fabrice Bellard’s vt100 for jslinux.
DugongCLEAN [menu]
Version of DugongGUI and DugongCMD without the Jupyter Notebook and Anaconda Navigator. It has a reduced size compared to other versions.
It has all the other tools available on Dugong.
Installing Bioinformatics Software [menu]
Two open-source projects were integrated with Dugong: BioConda and Linuxbrew, for the simple distribution of bioinformatics packages.
Conda and BioConda [menu]
Conda is a package manager that allows the installation, distribution and updating of different packages and their respective dependencies. It has a software distribution channel focused on bioinformatics tools called Bioconda.
Bioconda consists of:
- a repository of recipes hosted on GitHub
- a build system that turns these recipes into conda packages
- a repository of >2700 bioinformatics packages ready to use with conda install
- over 250 contributors that add, modify, update and maintain the recipes
For the installation of software through Conda or BioConda, simply execute the command below in a terminal in any version of Dugong:
conda install <software>
Browse packages in the BioConda channel: Available packages
For more details on the Conda Project, visit its documentation.
EXAMPLE: Installation in Dugong using BioConda:
- Video installation of bioinformatics tools: BWA and TOPHAT in DugongCMD. Installation performed through BioConda and operating test:
Anaconda Navigator [menu]
Anaconda Navigator is a desktop graphical user interface (GUI) that allows to launch applications and easily manage conda packages, environments and channels without using command-line commands.
Navigator is an easy, point-and-click way to work with packages and environments without needing to use conda commands in a terminal window. You can use it to find the packages you want, install them in an environment, run the packages and update them, all inside Navigator.
The DugongGUI version provides the Anaconda Navigator in its graphical interface with the Conda and BioConda repositories configured and functional.
Below is a preview of the Anaconda Navigator interface:
For more details on the Anaconda Navigator, visit its documentation.
EXAMPLE: Installation in Dugong using Anaconda Navigator:
- Video installation of bioinformatics tools: BWA and TOPHAT in DugongGUI. Installation performed through Anaconda Navigator and operational test:
LinuxBrew [menu]
Linuxbrew is a fork for Linux from Homebrew, the MacOS package manager, developed and maintained by Shaun Jackman. The project has Homebrew Science, a version focused on the distribution of scientific software.
For the installation of software of bioinformatics through Linuxbrew it is enough to execute the command below in a terminal in any version of Dugong:
brew install homebrew/science/<software>
or
brew tap homebrew/science
brew install <software>
Browse packages in the LinuxBrew channel: Available packages
For more details on the LinuxBrew Project, visit its documentation.
DEB Repository [menu]
In addition, it provides the BioLinux, CloudBioLinux and Linux Mint repositories for the apt-get package manager.
The installation of bioinformatics packages in Dugong is not restricted to the methods presented. Sources such as GitHub, Bitbucket, Sourceforge, Pypi, among others, can be used to install new tools simply and quickly.
Reproducibility and Replicability [menu]
Among the main objectives of the Dugong project is the provision of a complete work platform for the end user that allows the reproducibility and replicability of their research in the most diverse computational environments.
With this in mind, we implemented in all versions of Dugong the widely used Jupyter Notebook, among other utilities focused on the reproducibility of analyzes composed by complex computational environments and pipelines with different execution dependencies.
Jupyter Notebook [menu]
Jupyter Notebook extends the console-based approach to interactive computing in a qualitatively new direction, providing a web-based application suitable for capturing the whole computation analysis: developing, documenting, and executing code, as well as communicating the results. The Jupyter Notebook combines two components:
-
Web application: a browser-based tool for interactive authoring of documents which combine explanatory text, mathematics, computations and their rich media output.
-
Notebook documents: a representation of all content visible in the web application, including inputs and outputs of the computations, explanatory text, mathematics, images, and rich media representations of objects.
Main features of the web application:
- In-browser editing for code, with automatic syntax highlighting, indentation, and tab completion/introspection.
- The ability to execute code from the browser, with the results of computations attached to the code which generated them.
- Displaying the result of computation using rich media representations, such as HTML, LaTeX, PNG, SVG, etc. For example, publication-quality figures rendered by the matplotlib library, can be included inline.
- In-browser editing for rich text using the Markdown markup language, which can provide commentary for the code, is not limited to plain text.
- The ability to easily include mathematical notation within markdown cells using LaTeX, and rendered natively by MathJax.
Below is a preview of the Jupyter Notebook interface:

Dugong provided the Jupyter Notebook with Python 2.7.1, the IPython2 Kernel and the Notebook Conda Kernel, this extension allows administration of the entire Conda environment and a package installation through the Jupyter interface. Below is a preview of the Notebook Conda Kernel interface:
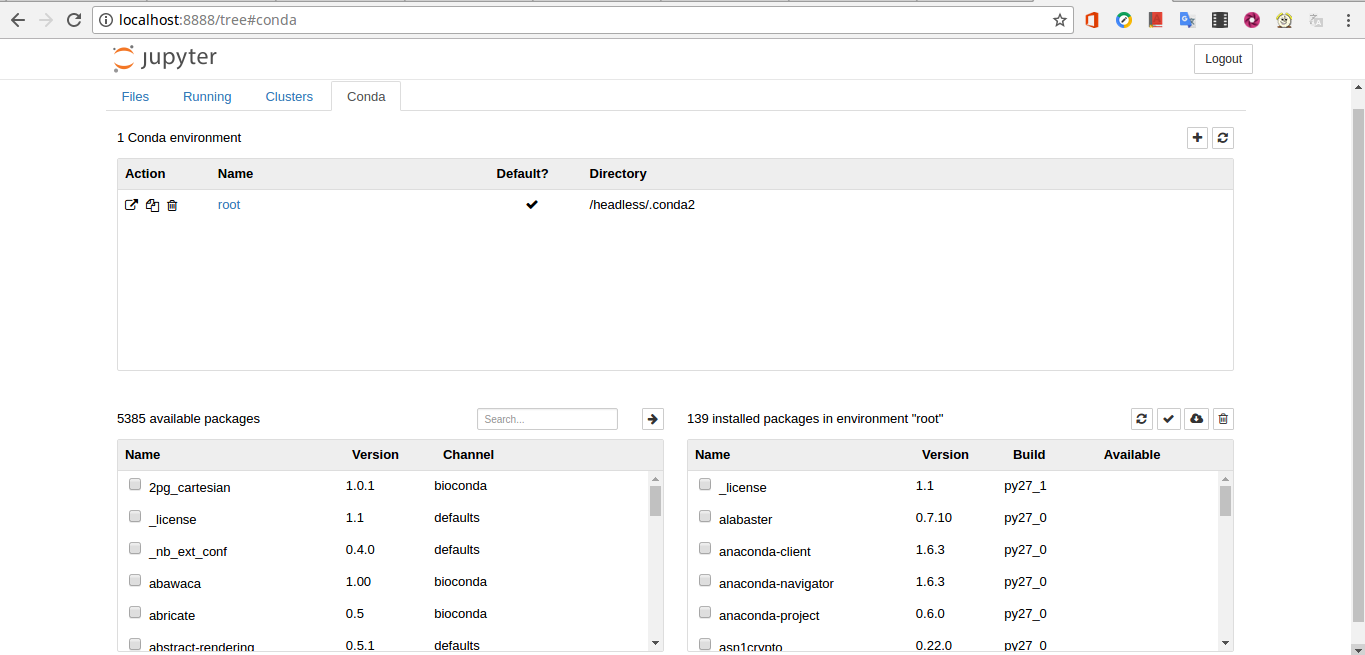
Getting the token and accessing the Jupyter Notebook [menu]
To access Jupyter Notebook, first deploy one of the versions of Dugong, according to our tutorials.
After deploy, enter the following command on the terminal of the host machine:
docker logs DugongGUI
or
docker logs DugongCMD
A message, similar to the one below, will be displayed:
[...]
[I 01:06:50.036 NotebookApp] [nb_conda_kernels] enabled, 2 kernels found
[W 01:06:50.258 NotebookApp] WARNING: The notebook server is listening on all IP addresses and not using encryption. This is not recommended.
[I 01:06:50.359 NotebookApp] [nb_conda] enabled
[I 01:06:50.365 NotebookApp] Serving notebooks from local directory: /headless/data
[I 01:06:50.365 NotebookApp] 0 active kernels
[I 01:06:50.365 NotebookApp] The Jupyter Notebook is running at: http://[all ip addresses on your system]:8888/?token=60a698e3d55cd77ffb61eb138e16fd874c11ab994199e859
[I 01:06:50.365 NotebookApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
[C 01:06:50.366 NotebookApp]
Copy/paste this URL into your browser when you connect for the first time,
to login with a token:
http://localhost:8888/?token=60a698e3d55cd77ffb61eb138e16fd874c11ab994199e859
Copy the Jupyter Notebook access address along with the token. Paste the address into the browser.
NOTE: If the access is not local, enter the IP / Hostname of the machine with the Jupyter Notebook next to the 8888 access port (Ex: http: // IP: 8888). In the new window, in the “Password or token” field, enter the previously obtained token.
For more details on the Jupyter Notebook, visit its documentation.
Script and ScriptReplay Commands [menu]
The script and scriptreplay commands are used to record and play terminal sessions on Linux distributions, respectively. The sum of these commands in a pipeline allows reproducibility and replicability at the Linux command line and is available in all versions of Dugong.
The script command stores terminal activities in a log file that will be named by a user, when a name is not provided by a user, the default file name is used. The scriptrepaly command helps to reproduce information in your log file used with the script command. The timing information is explained by the –timing = file option used with the command and the file generated by the previous command.
More details on the commands can be obtained through the video below:
Channel video: https://www.linuxhelp.com/script-scriptreplay-commands-linux-examples/
Deploy Dugong Image [menu]
Docker project provides a public cloud called the for sharing the developed containers. This cloud allows access to the application in a centralized and simple way, that is, it is possible to obtain a complete Dugong environment with command lines for its implementation.
To start a container, the user must have Docker installed on his operating system, according to the tutorials available in the project documentation. The Dugong image is available in the Docker Hub and its use is the recommended method of installation.
Two steps are required to start a container containing Dugong. In the first step, the Dugong image is downloaded from the Docker Hub servers to the host, and in the second, a container is created on the host machine with the default Dugong installation. If the host machine is a Linux, the following commands must be performed in the terminal:
docker pull dugong/dugong
docker run -d -p 5901:5901 -p 6901:6901 -p 8888:8888 -p 2222:22 --name Dugong -h Dugong -v $HOME/dugong/:$HOME/data/ \
--privileged dugong/dugong
At the end of the commands a Dugong instance will be available in the container named Dugong. Details about the container can be obtained through the command below:
docker ps
The default installation version of Dugong is DugongGUI with XFCE4.
To change the installed version simply add one of the tags in the deploy command: xfce4, icewm or cmd.
Deploy DugongGUI Image [menu]
Install DugongGUI Xfce4:
docker run -d -p 5901:5901 -p 6901:6901 -p 8888:8888 -p 2222:22 --name DugongGUI -h DugongGUI \
-v $HOME/dugongxfce/:$HOME/data/ --privileged dugong/dugong:xfce4
Install DugongGUI iceWM:
docker run -d -p 5901:5901 -p 6901:6901 -p 8888:8888 -p 2222:22 --name DugongGUI -h DugongGUI \
-v $HOME/dugongicewm/:$HOME/data/ --privileged dugong/dugong:icewm
EXAMPLE: Video installation DugongGUI:
- Click to watch the installation of DugongGUI on Linux Ubuntu Server:
- Click to watch the installation of DugongGUI on Linux CentOS 7 in the CloudatCost provider:
Deploy DugongCMD Image [menu]
Install DugongCMD:
docker run -d -p 3000:3000 -p 8888:8888 -p 2222:22 --name DugongCMD -h DugongCMD \
-v $HOME/dugongcmd/:$HOME/data/ --privileged dugong/dugong:cmd
EXAMPLE: Video installation DugongCMD:
- Click to watch the installation of DugongCMD on Linux Ubuntu Server:
- Click to watch the installation of DugongCMD on Linux CentOS 7 in the CloudatCost provider:
Deploy Dugong Image by graphical interface [menu]
Kitematic is a simple application for managing Docker containers on Mac and Windows, allowing the deploy of Docker images and the installation and administration of containers. Dugong can be installed through Kitematic or other softwares with the same function, such as Data Pulley.
Kitematic:

Data Pulley

For more details, visit the projects page.
Access Dugong Container [menu]
Access through the Docker console [menu]
Access to Dugong containers can be done in many ways, with access through the simplest Docker console. This access will not be of great attraction to the user without command line experience and should only be used in case of problems while running Dugong. Through this access, the user can restart Linux services, analyze system logs and applications, among other functions. To access the Docker console, simply run the following command on the Linux terminal on your host machine:
docker exec -it Dugong /bin/bash
Access through the SSH protocol [menu]
Access through the SSH protocol is also available in all versions of Dugong.
The Dugong execution command includes the “-p 2222: 22” option, where we direct the SSH port of the container to port 2222 of the host. To access Dugong by SSH, from your host machine, simply execute the following command:
ssh -p 2222 dugong@localhost
The dugong user’s default password (dugong) will be requested at that time. This password can be changed through the command: passwd dugong
Access through the VNC and noVNC [menu]
Two other methods for accessing DugongGUI is through VNC or noVNC. Ports 6901 and 5901 are respectively the VNC and noVNC execution ports. These ports are specified during container creation on the host machine and can be changed as per Docker documentation (-p 6901:6901 -p 5901:5901).
To access noVNC it is enough that the user directs his navigator to the address below, after starting the Dugong container. The default password for noVNC access is vncpassword and can be changed at any time. The default DugongGUI user is the dugong with the dugong password.
http://<IP or Host>:<port>/vnc_auto.html?password=vncpassword
A client is required for Dugong access through the VNC protocol. During the tests, the VNC® Viewer for Google Chrome application was used in the Chromebook:
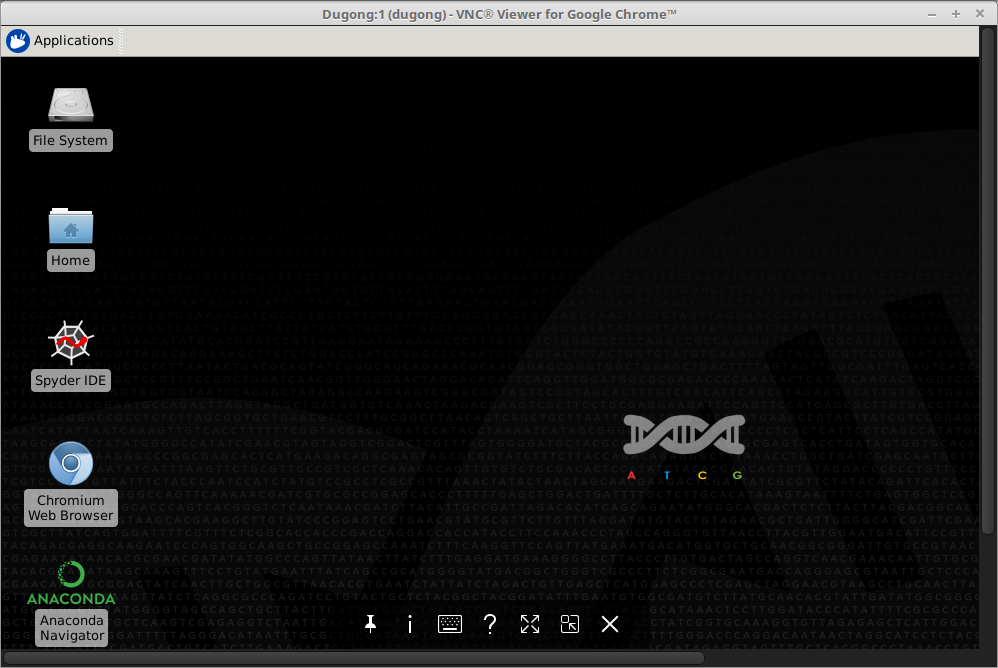
Access through a Web Terminal in Node.js [menu]
For access to DugongCMD the user can use an SSH client of his choice. We also provide access through the tty application, requiring only that the user direct their browser to the address below followed by port 3000. The default DugongCMD user is the dugong with the dugong password.
http://<IP or Host>:3000
Example of access through the Web Terminal in a Google Chrome:
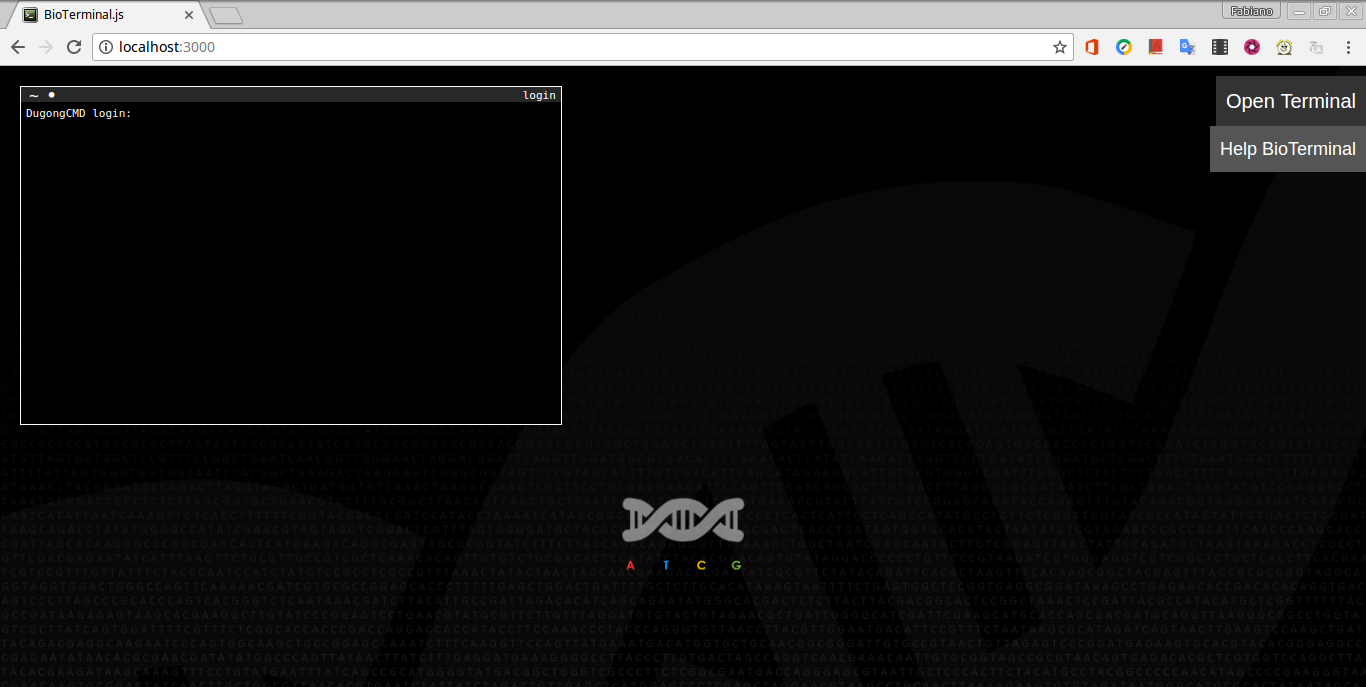
Extending or adapting the Dugong image [menu]
Dugong can be expanded or adapted to the most diverse needs in a research or teaching environment. All Dugong environment configuration scripts are available in the Git Hub (https://dugongbioinformatics.github.io/), including Dockerfile for building your image.
Example of adapted tools, replicability and reproducibility in Dugong [menu]
CirComPara Pipeline [menu]
CirComPara is a computational pipeline to detect, quantify, and correlate expression of linear and circular RNAs from RNA-seq data.
- Video from a CircRNA analysis using the CirComPara tool on an Ubuntu Server with a DugongGUI container installed. The video presents a complete implementation of CirComPara with the test data provided by the authors and their expected results:
For more information on adapting CirComPara to Dugong and accessing analysis results using Dugong and CirComPara in different computing environments, go to: DugongGUI CirComPara
Dugong Virtual Box [menu]
The Dugong Virtual Box gets around the difficulty of installation by providing a functioning Dugong full install inside an Ubuntu, Fedora or Arch Linux virtual machine. You can use the Dugong Virtual Box on Mac OS X, Windows, or Linux.
Deploy Dugong Virtual Box [menu]
It is strongly recommended that your system have 8 gigabytes or more of memory to use the Dugong Virtual Box.
- Download and install the VirtualBox (VB) version for your machine.
- Download the 64-bit Dugong Virtual Box, which is linked from the Dugong Resources page. This file is large so it may take between a few minutes and a few hours depending on your Internet connection speed. You will need to unzip this file, which you can typically do by double-clicking on it.
- In the File menu, select Import Appliance. The Appliance Import wizard is displayed in a new window.
- Click Choose, browse to the location containing the *.ovf or *.ova file of the virtual machine of Dugong, and click Open.
- Make any adjustments you want to the displayed settings (you can also change the settings later) and click Import. The Appliance Import Wizard is closed and after a few moments, the imported virtual machine is listed in Oracle VM VirtualBox Manager.
- After the import, select the imported virtual machine and in the toolbar click the Settings button. Review the virtual machine settings to make sure that the virtual machine has the hardware it needs to operate. Make sure that the virtual machine has a CD/DVD drive.
- Once you have reviewed the settings, select the imported virtual machine and in the toolbar click the Start button. Verify that the virtual machine works.
-
After the start of your virtual machine, execute the command below:
docker start Dugong docker start DugongCMD docker start Dugong-CirComPara -
After starting the containers, enter the command below to obtain the ports for access through a browser:
docker ps -
Finally, access the addresses below through a browser to access the respective versions of Dugong:
DugongGUI and CirComPara: http://localhost:<port>/vnc_auto.html?password=vncpassword (ports displayed in the above command) DugongCMD: http://localhost:3000 (login: dugong / password: dugong)
Comparison between Dugong and other bioinformatics services [menu]
We provide a comparison of Dugong with the main tools available for analysis of bioinformatics in virtualized platforms and Docker.
To view this comparison click here.
Author [menu]
Current development is led by Fabiano Menegidio.
How to cite [menu]
Menegidio, F.B. et al. (2018). Dugong: a Docker image, based on Ubuntu Linux, focused on reproducibility and replicability for bioinformatics analyses. Bioinformatics 34(3):514-515. doi: 10.1093/bioinformatics/btx554.
Contributing [menu]
You are invited to contribute new features, fixes, or updates, large or small; we are always thrilled to receive pull requests, and do our best to process them as fast as we can.
Before you start to code, we recommend discussing your plans through a GitHub issue, especially for more ambitious contributions. This gives other contributors a chance to point you in the right direction, give you feedback on your design, and help you find out if someone else is working on the same thing.
MIT License [menu]
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
The software is provided “as is”, without warranty of any kind, express or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the authors or copyright holders be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the software or the use or other dealings in the software.
Learn More [menu]
To learn more about Docker, go to:
http://docker.com
https://docs.docker.com/